Supercharge Your Data Workflow with Custom Slash Commands in Gemini CLI
Turn your repetitive data steps into one-line AI-powered commands.
I have been working in data space for almost 10 years and there is one thing that never changed. Every time I receive a dataset (CSV, Parquet, etc), the routine is always the same:
fire up my python or jupyter notebook, import pandas (or now polars), run df.head(), check the dataset shape, run df.info(), df.describe() and only then I can start the actual work.
Anyone working with data, whether you are a data engineer, analyst or scientist knows this routine. It’s mundane and repetitive. And honestly, it’s something that I should have automated years ago.
Then I stumbled onto Gemini CLI’s custom slash commands and it’s all clicked.
What this article is about?
This is the first post in a series where I will share the custom commands I have built to streamline my data workflow. In this post, I will walk through the first custom slash command: /preview. This command gives me quick peek at the dataset from the terminal.
By the end of this post, you will understand:
How the
/previewcommand worksThe design behind it
Why I chose a hybrid approach (shell commands + LLM)
How to build your own version
Disclaimer: I am experimenting on these commands and I’ll be sharing more as my workflow improves. It’s a work in progress that actually works.
The problem: previewing datasets is boring, but necessary
Here is a example of usual routine in pandas:
import pandas as pd
df = pd.read_csv(’users.csv’)
print(df.head())
print(df.shape)
print(df.info())This works, but it also means firing up python, waiting for pandas to load, typing same four lines over and over, and sometimes switching between windows — you get the idea.
Enter /preview: my shortcut for data preview
As the name suggests, it instantly previews your dataset (for now only CSVs), doing it smarter, faster and far less repetitively than the manual approach above.
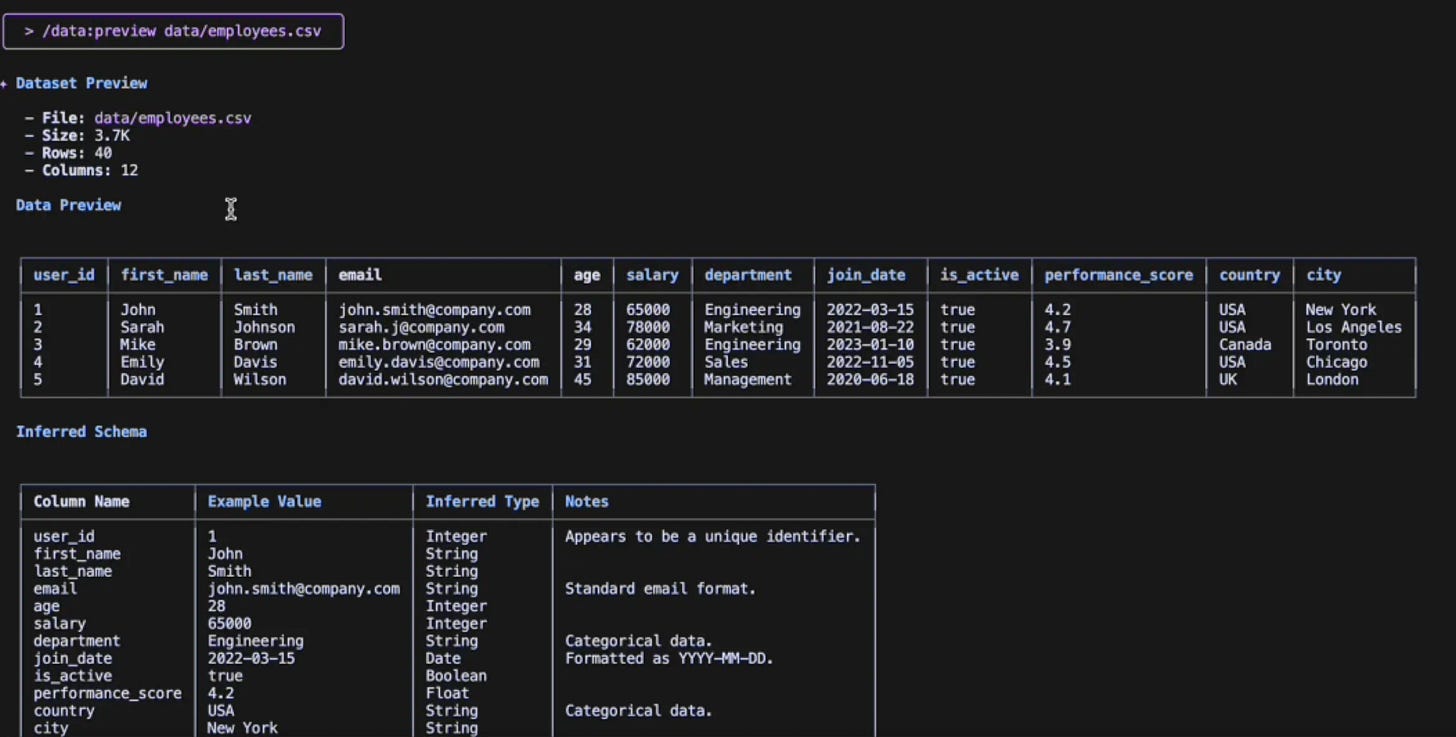
/preview data/employees.csvWith this single command on Gemini CLI, I get:
The first 5 rows in a markdown table
The inferred schema with data types
The dataset dimensions (rows and columns)
And that’s not all, this command will also perform validations to check whether the file actually exists and if it’s a valid CSV file.
How it works: the anatomy of preview.toml
I will now discuss how I built this command. The full preview.toml is at the end, but I want to break down and explain the key pieces first.
Part 1: the setup
description = “Previews the first 5 rows of a CSV file and shows total rows and columns and the inferred schema.”This is what shows up when you run /help in Gemini CLI. Keep it concise so you (and future you) know what it does at a glance.
Part 2: guardrails (file validation)
First things first, it’s important to verify that the file exists and is readable.
File validation:
!{[ -f “{{args}}” ] && echo “✓ File exists” || echo “✗ File not found”}
File info:
!{ls -lh “{{args}}” 2>/dev/null || echo “Cannot access file”}The !{...} syntax tells Gemini CLI to run a shell command. Here I use it to:
Confirm the file exists (
[ -f “{{args}}” ])Show the file size (
ls -lh)Handle the errors gracefully
With these validations I can catch problems upfront rather than dealing with confusing errors later.
Part 3: data extraction (the shell commands)
Here’s the fun part. Rather than making the LLM read and parse the entire CSV (slow an memory-heavy ), I use the shell commands to pull just what I need:
Total rows (excluding header):
!{if [ -f “{{args}}” ]; then tail -n +2 “{{args}}” | wc -l | tr -d ‘ ‘; else echo “N/A”; fi}
Column count (first row):
!{if [ -f “{{args}}” ]; then head -n 1 “{{args}}” | awk -F’,’ ‘{print NF}’ | tr -d ‘ ‘; else echo “N/A”; fi}
Data preview (header + 5 rows):
!{if [ -f “{{args}}” ]; then head -n 6 “{{args}}”; else echo “No data available”; fi}Here’s the breakdown:
tail -n +2 | wc -l: skip the header, count the rowshead -n 1 | awk -F’,’ ‘{print NF}’: get the first row, count columnshead -n 6: grab the header + 5 data rows
Part 4: LLM does its thing
The shell commands provide us with raw data, and now we ask the LLM to interpret it:
**Required Output Format:**
# Dataset Preview
- **File:** `{{args}}`
- **Size:** [Extract file size from ls output above]
- **Rows:** [Extract row count from above, excluding header]
- **Columns:** [Extract column count from above]
## Data Preview
[Convert the CSV preview data above into a clean Markdown table...]
## Inferred Schema
[Analyze the preview data and create a table with: | Column Name | Example Value | Inferred Type | Notes |In this case, the LLM will do the following:
Show dataset details such as file name, size, total rows and colums
Format the data into clean markdown tables
Infer data types (String, Integer, Float, Date, etc.)
This is what LLMs excel at: spotting the patterns, formatting neatly and giving context. Counting rows? That’s the shell’s job. Reading files? Handled by the OS. The LLM’s role is to make sense of it all.
Why using hybrid approach? why not just use the LLM?
I initially tried using the LLM for the entire task, but reading and parsing a full dataset takes time. And for the /preview command, I don’t actually need the whole file. Just specific pieces of information where I can use the shell commands to extract them.
And in addition to that, why burn tokens (cost) for counting rows when wc -l can do the job in microseconds? This makes it much more efficient & practical.
The hybrid approach plays to each tool’s strengths:
Shell: Fast file operations, data extraction
LLM: Pattern recognition, schema inference, insights
The full code
Here is the complete preview.toml file:
description = “Previews the first 5 rows of a CSV file and shows total rows and columns and the inferred schema.”
prompt = “”“
You are a senior data analyst. You will analyze CSV file information and present it in a structured format.
**Instructions:**
1. Validate that the provided file path exists and appears to be a CSV file
2. If there are any errors (file not found, not readable, etc.), respond with a clear error message
3. Otherwise, format the information according to the specified template below
**File Path:** {{args}}
**Shell Commands Output:**
```
File validation:
!{[ -f “{{args}}” ] && echo “✓ File exists” || echo “✗ File not found”}
File info:
!{ls -lh “{{args}}” 2>/dev/null || echo “Cannot access file”}
Total rows (excluding header):
!{if [ -f “{{args}}” ]; then tail -n +2 “{{args}}” | wc -l | tr -d ‘ ‘; else echo “N/A”; fi}
Column count (first row):
!{if [ -f “{{args}}” ]; then head -n 1 “{{args}}” | awk -F’,’ ‘{print NF}’ | tr -d ‘ ‘; else echo “N/A”; fi}
Data preview (header + 5 rows):
!{if [ -f “{{args}}” ]; then head -n 6 “{{args}}”; else echo “No data available”; fi}
```
**Required Output Format:**
# Dataset Preview
- **File:** `{{args}}`
- **Size:** [Extract file size from ls output above]
- **Rows:** [Extract row count from above, excluding header]
- **Columns:** [Extract column count from above]
## Data Preview
[Convert the CSV preview data above into a clean Markdown table. If the file doesn’t exist or has errors, show the error message instead]
## Inferred Schema
[Analyze the preview data and create a table with: | Column Name | Example Value | Inferred Type | Notes |
For “Inferred Type”, use: String, Integer, Float, Boolean, Date, or Mixed
For “Notes”, provide brief insights about the data pattern, nulls, or formatting]
**Important:**
- If any shell command failed or the file doesn’t exist, start your response with “❌ **Error:**” and explain the issue
- Only proceed with the full analysis if all validations pass
“”“To install it:
# Create the commands directory if it doesn’t exist
mkdir -p ~/.gemini/commands
# Create the file
touch ~/.gemini/commands/preview.toml
# Paste the code aboveThen just run this command on your Gemini CLI:
/preview <file_path_to_your_data.csv>What’s next
This is just the first command in what’s becoming a full toolkit. I am currently working on:
/describe: statistical summaries (thinkdf.describe())/dq-report: for data quality checks/transform: common data cleaning operations/compare: diff between two CSV files
If you want to build your own commands, check out the Gemini CLI commands documentation and this tutorial by Romin Irani. The custom command system is surprisingly powerful once you understand the basics.
Final thoughts
I am not saying this is revolutionary or groundbreaking. It’s really just a couple of shell commands inside a TOML file that talks to an LLM. And honestly, that’s enough for me. Automation doesn not have to be flashy or fancy. If it saves me time, I will happily use it.
Drop a comment. I would love to hear your feedback and learn how others are using custom slash commands to automate their data workflows. Thank you for reading and Let’s get connected!